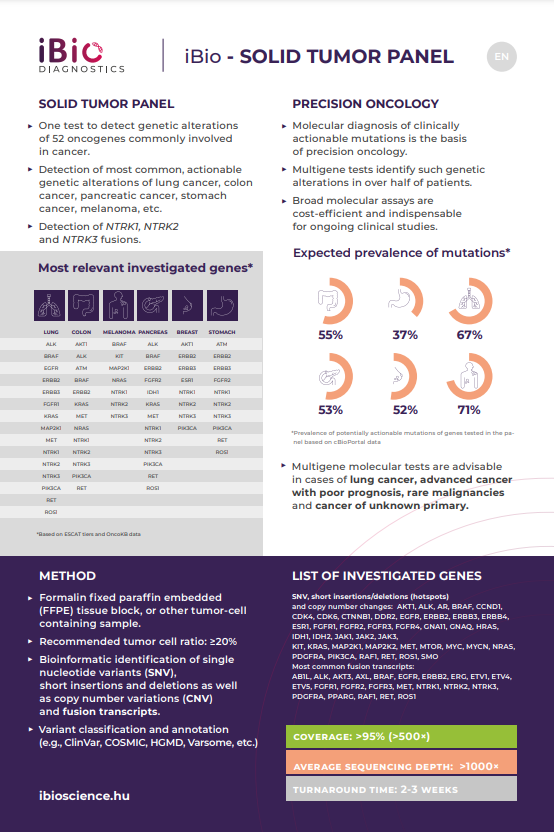
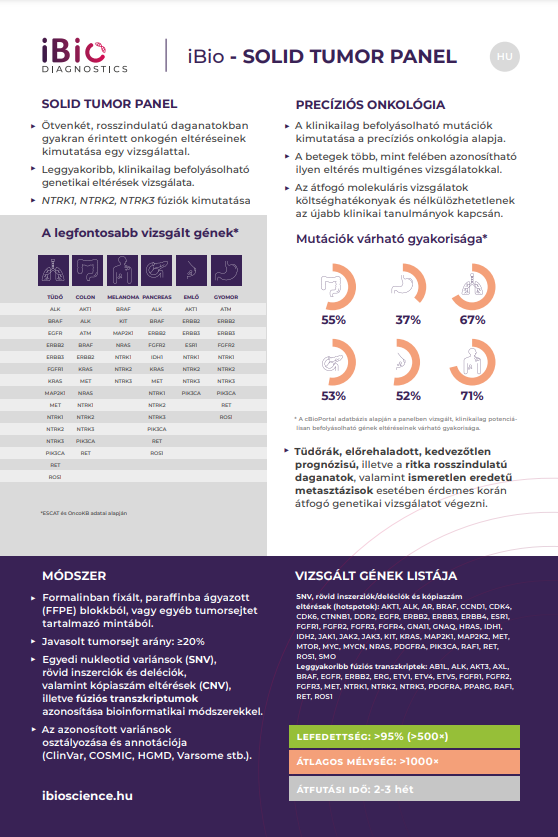
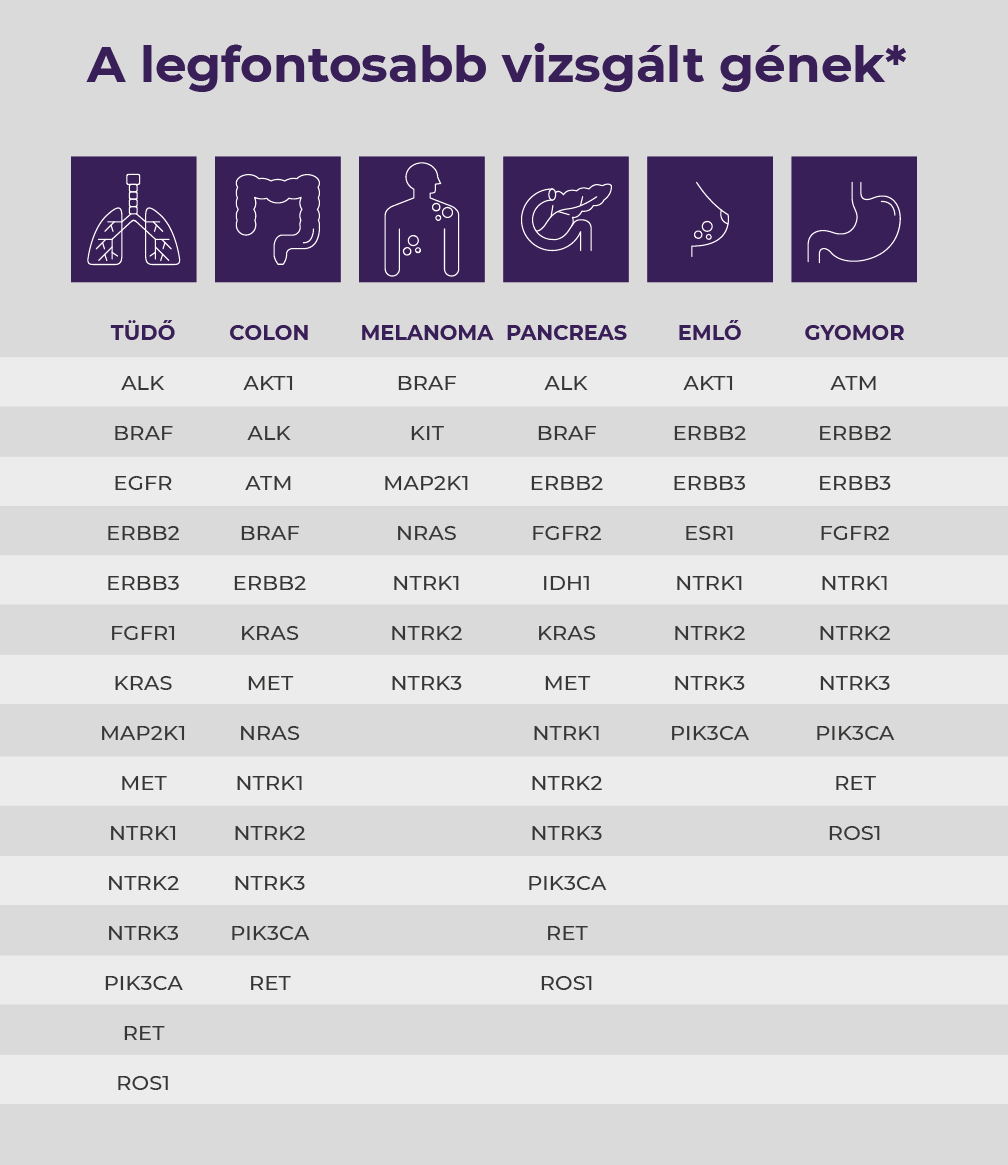
- Detection of fifty-two oncogenic abnormalities commonly involved in malignancies in one test.
- Examination of the most common genetic abnormalities that can be clinically influenced.
- Detection of NTRK1, NTRK2, NTRK3 fusions.
Precision oncology
- The detection of clinically actionable mutations is the basis of precision oncology.
- In more than half of patients, such abnormalities can be identified by multigene testing.
- Comprehensive molecular studies are cost-effective and essential for new clinical studies.
- For lung cancer, advanced tumours with an unfavourable prognosis, rare malignancies and metastases of unknown origin, it is worthwhile to perform comprehensive genetic testing early.
Expected frequency of mutations

CONTACT
+36 70 674 6611

PROJECT ORGANIZATION
project consultation, software demo, price quote, contract, organization

SAMPLE SUBMISSION
FFPE block or section, cell block, or smear, or isolated DNA

RESULT REPORTING
classification of variants according to international standards (pathogenic, likely pathogenic, VUS, etc.)
GENETIC ANALYSIS
bioinformatics analysis, identification and annotation of variants

HISTOLOGICAL PROCESSING
microscopic assessment of tumor cell number, DNA isolation
Method
- From a formalin-fixed, paraffin-embedded (FFPE) block or other tumour cell-containing specimen.
- Recommended tumour cell percentage: ≥20%
- Identification of single nucleotide variants (SNVs), short insertions and deletions, copy number variations (CNVs) and fusion transcripts using bioinformatics methods.
- Classification and annotation of identified variants (ClinVar, COSMIC, HGMD, Varsome, etc.).
List of genes tested
SNV, short insertions/deletions and copy number discrepancies (hotspots): AKT1, ALK, AR, BRAF, CCND1, CDK4, CDK6, CTNNB1, DDR2, EGFR, ERBB2, ERBB3, ERBB4, ESR1, FGFR1, FGFR2, FGFR3, FGFR4, GNA11, GNAQ, HRAS, IDH1, IDH2, JAK1, JAK2, JAK3, KIT, KRAS, MAP2K1, MAP2K2, MET, MTOR, MYC, MYCN, NRAS, PDGFRA, PIK3CA, RAF1, RET, ROS1, SMO Most common fusion transcripts: AB1L, ALK, AKT3, AXL, BRAF, EGFR, ERBB2, ERG, ETV1, ETV4, ETV5, FGFR1, FGFR2, FGFR3, MET, NTRK1, NTRK2, NTRK3, PDGFRA, PPARG, RAF1, RET, ROS1
COVERAGE
>95% (>500×)
AVERAGE DEPTH
>1000×
TURNAROUND TIME
2-3 hét