- Classification and annotation of identified variants (ClinVar, COSMIC, HGMD, Varsome, etc.)
- Classification and annotation of identified variants (ClinVar, COSMIC, HGMD, Varsome, etc.)
- Malignant diseases are about. 50% have a mismatch in the TP53 gene.
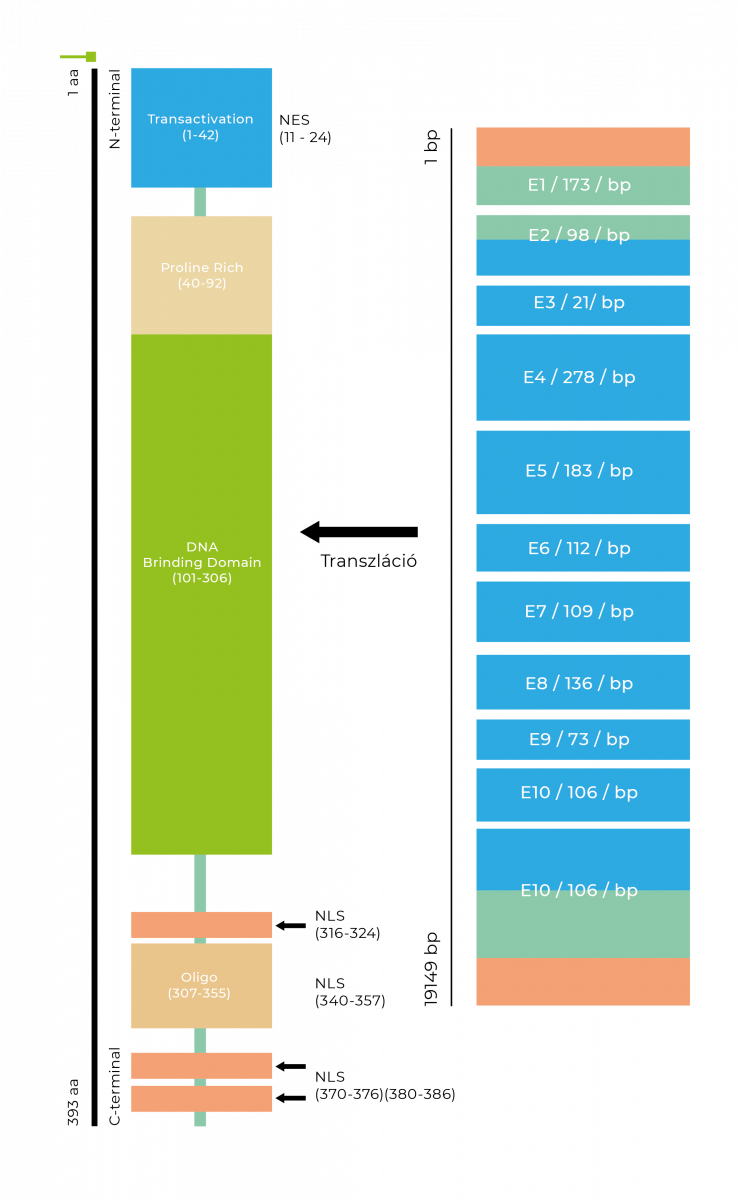
- Pathogenic mutations can occur in all 11 exons of the 32.8 kb gene.
- The iwCLL (International Working Group on CLL) recommends TP53 mutation analysis before therapy for all patients with chronic lymphocytic leukaemia.
Chronic lymphocytic leukaemia
- Chronic lymphocytic leukaemia (CLL) is the most common adult leukaemia and is a genetically heterogeneous disease.
- At diagnosis, TP53 mutations can be detected in 4-7% of patients, but in therapy-refractory cases this rate can be as high as 40-50%.
- The TP53 mutation is the most important prognostic and predictive genetic abnormality in CLL; it is associated with an unfavourable disease course and failure of chemoimmunotherapy.
- The iwCLL (International Working Group on CLL) recommends TP53 mutation analysis before therapy for all patients with chronic lymphocytic leukaemia.
Acute leukaemias
- Acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL) are poor prognosis, highly malignant, genetically heterogeneous diseases in adults.
- TP53 abnormalities occur in 5-15% of acute leukaemias and are associated with an unfavourable disease course.
- TP53 mutation analysis is recommended for all AML and ALL.

CONTACT
+36 70 674 6611

PROJECT ORGANIZATION
project consultation, software demo, price quote, contract, organization

SAMPLE SUBMISSION
FFPE block or section, cell block, or smear, or isolated DNA

RESULT REPORTING
classification of variants according to international standards (pathogenic, likely pathogenic, VUS, etc.)
GENETIC ANALYSIS
bioinformatics analysis, identification and annotation of variants

HISTOLOGICAL PROCESSING
microscopic assessment of tumor cell number, DNA isolation
Method
- From isolated genomic DNA or EDTA bone marrow aspirate/peripheral blood sample, if necessary from a paraffin-embedded (FFPE) block fixed in formalin.
- Szükséges tumorsejt arány: ≥20%
- Sequencing of the entire coding and 3'/5' UTR regions of the TP53 gene.
- Bioinformatic identification of single nucleotide variants (SNV), short insertions and deletions.
- Classification and annotation of identified variants (ClinVar, COSMIC, HGMD, IARC TP53, etc.).
COVERAGE
>95% (>500×)
AVERAGE DEPTH
>1000×
TURNAROUND TIME
2-3 hét